Help for Contaminated Groundwater: Dioxane-chomping Microbe
Published on by Water Network Research, Official research team of The Water Network in Academic
Rice University researchers have discovered a bacteria-borne gene that helps degrade a form of dioxane, a groundwater contaminant and suspected carcinogen.
The discovery could be the basis for a much-needed tool to decide how contaminated sites should be treated.
Source: ACS
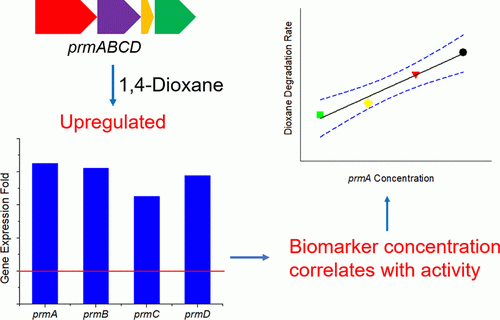
Research by the Rice lab of civil and environmental engineer Pedro Alvarez found a novel gene cluster in Mycobacterium dioxanotrophicus PH-06, a mold-like bacterium capable of using 1,4-dioxane as its sole source of carbon and energy. The microbe was discovered in 2009 in the sediment of a dioxane-contaminated river in South Korea.
In an earlier study, the Rice team decoded the complete genome sequence of the bacterium. While it clearly fed on dioxane, it did not contain a well-studied gene found in another microbe known to initiate dioxane biodegradation.
But the whole genome sequence revealed the presence of a novel propane mono-oxygenase gene cluster that expresses an enzyme the researchers said is also likely to initiate dioxane biodegradation. “This is important because it shows that dioxane-degrading genes are more diverse than previously appreciated,” Alvarez said.
The new study appears in the American Chemical Society journal Environmental Science & Technology Letters.
Dioxane has been widely used as a stabilizer for chlorinated solvents and is commonly found as a co-contaminant at thousands of polluted sites, according to the researchers. The chemical is highly soluble in water, easily leaches into groundwater and is resistant to natural biodegradation, Alvarez said.
He said the discovery should help environmental engineers find the best way to treat contaminated groundwater.
“Current site remediation approaches like ‘pump and treat’ or in situ chemical oxidation are not suitable for large and dilute dioxane plumes that prevail at thousands of contaminated sites,” he said. “For such plumes, monitored natural attenuation (MNA) can be the most cost-effective approach.
“However, MNA is underutilized at dioxane-impacted sites because we lack reliable analytical tools to support decisions to select it or reject it,” he said.
The gene cluster could be employed as a probe to test for the presence of dioxane degraders in groundwater. If degraders are present, the site might be best left to eliminate dioxane naturally, though with continued monitoring. The gene cluster provides a basis for minimizing false negatives when looking for dioxane degraders at contaminated sites, Alvarez said.

Representative image, Source: Max Pixel
“There’s an urgent need for these molecular tools from an economic standpoint,” he said. “Our novel gene probe can enlighten judicious MNA selection and avoid significant expenses associated with costly and marginally effective remediation alternatives.”
Rice graduate student Ya He is lead author of the study. Co-authors are research scientists Jacques Mathieu and Márcio Luís Busi Da Silva and postdoctoral researchers Yu Yang and Pingfeng Yu. Alvarez is the George R. Brown Professor in civil and environmental engineering and in materials science and nanoengineering and director of the Rice-based, National Science Foundation-sponsored Nanosystems Engineering Research Center for Nanotechnology-Enabled Water Treatment.
The Department of Defense’s Strategic Environmental Research and Development Program sponsored the research.
Read the abstract at ACS
Source: Rice
Media
Taxonomy
- Quality
- Water Quality
- Groundwater
- Hydrodynamics & Water Quality
- Pollution
- Groundwater Assessment
- Groundwater Pollution
- Groundwater Quality & Quantity
- Pollution